Phase 3 Questions & Answers
This page records specific questions of Phase 3 users and the corresponding answers given by the ESO/ASG support team assuming that the provided information appears to be helpful to a broader audience.
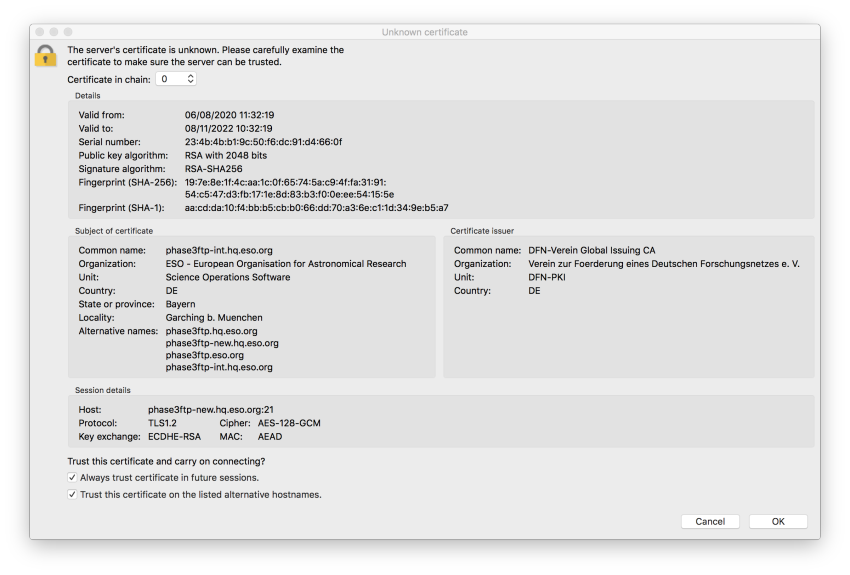
FileZilla: unknown certificate error
Q: In trying to upload my files I encountered an error message, how can I proceed?
The error is caused by FileZilla not trusting the OS certificate stack. In order to proceed, you have to confirm explicitly that the server can be trusted. Having verified that the server’s fingerprint and the certificate fingerprint displayed in the figure are identical, please accept the server certificate by checking the two options as shown in the figure below.
Renaming downloaded Phase 3 products to their original file names
Q: How can I rename the P3 products from their ADP names to the original file names?
A: In the case of files downloaded via the Request Handler, the correspondence between file-id and original file-name is mapped in the README file associated to each data request. The renaming commands can be extracted directly from the README file using the following shall command:
awk '/\- ADP/{ print "mv "$2, $3}' README_123456.txt
In some cases it can happen that during the downloading the ':' of the file name is replaced by a '_', if so, please use this other command line:
awk '/\- ADP/{ print "mv "$2, $3}' README_123456.txt | awk '{gsub(/\:/, "_", $2)}1'
Migrating catalogues from ASCII to FITS format
Q: How to convert an ascii file (table) to a fits file?
A: You can perform the following steps:
- Create a comma separated ascii table. For example if the table (cat1.txt) has n columns separated by white spaces:
awk '{print $1,","$2,","$3,","$...,","$n-1,","$n}' cat1.txt > cat2.txt - Convert the new table (cat2.txt) into a fits file using the stilts tool:
stilts tcopy cat2.txt cat.fits ifmt=csv ofmt=fits-basic
Usage:
stilts <stilts-flags> tcopy ifmt=<in-format> ofmt=<out-format> [in=]<table> [out=]<out-table>
- Add the missing header keywords according to the ESO Science Data Products Standard.
The STIL Tool Set can be downloaded from:
http://www.star.bris.ac.uk/~mbt/stilts/#install
For more information please check the following documentation:
http://www.star.bris.ac.uk/~mbt/stilts/
http://www.star.bris.ac.uk/~mbt/stilts/sun256/tcopy.html
Producing Phase 3 compliant spectra
Q: How can I convert my spectrum to the binary table format required by the Phase 3 Standard?
A: You can use the following Python script (requirements: Python-2.7 or greater, astropy library) but please make sure to modify it by providing the correct values for the required FITS header keywords to be added to the file. To help the users the script already includes the commands to add all the Phase 3 mandatory keywords but when the value must be inserted by the user, the line is commented. Please provide the correct value and de-comment the line in order to generate a Phase 3 complaint file.
- In case the original spectrum is an image with the wavelength in the WCS, use this script. You can give it a try by using this input file. The script should output the following converted spectrum.
Preparing APEX 1-D spectra compliant to the Phase3 standard
Q: Could you please provide some information regarding the format of APEX 1-D spectra?
A: The format defined in the Phase 3 SDPS for 1-D spectral products applies also in the case of APEX reduced spectra, with few modifications:
1. The following keyword defined as mandatory for spectral products do not apply in the case of APEX spectra: OBID1.
2. While the keyword FEBE1 indicating the Frontend-backend combination becomes mandatory in this case.
Example of the values of some specific keywords:
Primary header
ORIGIN = 'APEX' / Facility TELSCOP = 'APEX-12m' / Telescope name INSTRUME= 'APEXHET' / Instrument name FEBE1 = 'HET230-XFFTS2' / Frontend-backend combination OBSTECH = 'SPECTRUM' / Technique of observation PRODCATG= 'SCIENCE.SPECTRUM' / Data product category
Extension header
EXTNAME = 'SPECTRUM' / FITS Extension name TTYPE1 = 'FREQ ' / Label for field 1 TUTYP1 = 'Spectrum.Data.SpectralAxis.Value' TUNIT1 = 'GHz ' / Physical unit of field 1 TUCD1 = 'em.freq ' / UCD of field 1 TDMIN1 = 229.011 / Start in spectral coord. TDMAX1 = 233.000 / Stop in spectral coord. TTYPE2 = 'FLUX ' / Label for field 2 TUTYP2 = 'Spectrum.Data.FluxAxis.Value' TUNIT2 = 'mJy ' / Physical unit of field 2 TUCD2 = 'phot.flux.density;em.freq' / UCD of field 2 TTYPE3 = 'ERR ' / Label for field 3 TUTYP3 = 'Spectrum.Data.FluxAxis.Accuracy.StatError' TUNIT3 = 'mJy ' / Physical unit of field 3 TUCD3 = 'stat.error;phot.flux.density;em.freq' / UCD of field 3
Please also note that the values of the following keywords need to be provided in nm:
WAVELMIN= 1291867.18750 / [nm] Minimum wavelength
It corresponds to the value of TDMIN1 converted from GHz to nm (conversion factor: 299792458)
WAVELMAX= 1309075.80566 / [nm] Maximum wavelength
It corresponds to the value of TDMAX1 converted from GHz to nm (conversion factor: 299792458)
SPEC_BIN= 111.744273793 / [nm] Wavelength bin size SPEC_VAL= 1300471.49658 / [nm] Mean Wavelength SPEC_BW = 17208.6181641 / [nm] Bandpass Width Wmax – Wmin
The following link may be useful to reconstruct the provenance information, it provides the mapping between the original file name to the archive id, and calculating the exposure time:
http://archive.eso.org/wdb/wdb/eso/apex_origfile/form
Specification of keywords for data originating from non-ESO telescopes
Possible algorithm for ABMAGSAT computation
Q: Possible algorithm for ABMAGSAT computation
In the case of the VIMOS imaging pipeline, an adaptation of the method used by the PESSTO survey (A&A, 2015, 579, pg. 25) is the following:
ABMAGSAT = zeropoint -2.5*log10(((pi/4.*ln(2)))*(satlev-mean_sky)*(psf_fwhm/pixel_scale)^2)/EFF_EXPT)
where the parameters are those written to the following keywords:
satlev = HIERARCH ESO QC SATURATION zeropoint = HIERARCH ESO QC MAGZPT mean_sky = HIERARCH ESO QC MEAN_SKY pdf_fwhm = PSF_FWHM pixel_scale = HIERARCH ESO QC WCS_SCALE EFF_EXPT = the effective exposure time (= CASUEXPT)
Computing MJD-END of SOFI spectra
Q: How to compute the MJD-END of a SOFI spectrum?
The end time of a Phase 3 SOFI 1d spectrum product (MJD-END) must be computed using the following formula:
MJD-END = MJD-OBS of the last raw observation + NDIT * ( DIT + 1.8) / 86400
where the 1.8 seconds accounts for the necessary overheads, and 86400 scales back from seconds to days.
EFFRON for median-combined SOFI images
Q: What is the correct EFFRON for median-combined SOFI images?
Example: There are 7 raw images, each resulting from averaging together 5 detector integrations (NDIT = 5). A science product is generated by reducing and median-combining those 7 raw images.
In this case:
EFFRON = 12 * sqrt(PI/2) / sqrt( 7 * 5)
where PI is 3.14159, and 12 is the detector readout noise of SOFI in electrons.
OBSTECH values
Q: May you please clarify what the OBSTECH keyword values are?
A: We support the OBSTECH keywords listed in the table below, in addition to those defined by the SDP standard document.
| INSTRUME |
Mode | Origin of keyword value | TELESCOP | OBSTECH |
|
OSIRIS
|
Imaging | Broad band (SDSS filters) | OBSMODE | GTC | 'IMAGE' 'IMAGE,FABRY-PEROT' 'IMAGE' |
| Narrow band (Tunable filters) | |||||
| Medium band (SHARDS filters) | |||||
| Spectroscopy | Long slit | 'SPECTRUM' | |||
| CanariCAM | Imaging | 'IMAGE,CHOPNOD' 'IMAGE,NODDING' 'IMAGE,CHOPPING' 'IMAGE,STARE' | |||
| Spectroscopy | 'SPECTRUM,CHOPNOD' 'SPECTRUM,NODDING' 'SPECTRUM,CHOPPING' 'SPECTRUM,STARE' | ||||
